Llama_index Source Code Analysis(1)
Summary
This blog introduces the basic concepts of RAG and further demonstrates the RAG process based on the source code interpretation of llama_index, including data loader, transformation, index, query, etc. In addition, this paper also analyzes the performance of llama_index RAG process and gives corresponding optimization suggestions.
1. Introduction
Llama_index is a framework designed to build context-enhanced large model applications. It leverages private user data to improve model performance in specific domains.
Llama_index primarily offers the following tools:
- Data Connector: Connects to private user data, APIs, databases, etc.
- Data Indexes: Structures data in a format conducive to large language models (LLMs).
- Engines: Provides natural language access methods:
- Query Engine: Interfaces for question-answering, such as knowledge base queries.
- Chat Engine: Interfaces for multi-turn dialogues, like GPT.
- Agents: Services based on LLMs, such as task automation, customer service, etc.
- Observability/Evaluation: Integrates tools for application evaluation and monitoring.
This analysis is based on version llama-index==0.10.40.
2. RAG High Level Concepts
RAG stands for Retrieval-Augmented Generation.
Typically, large models are trained on public datasets, but they may perform suboptimally for specific tasks. RAG incorporates private user data into the accessible data for the model, feeding it as context along with the query. This process does not require fine-tuning or training the model.
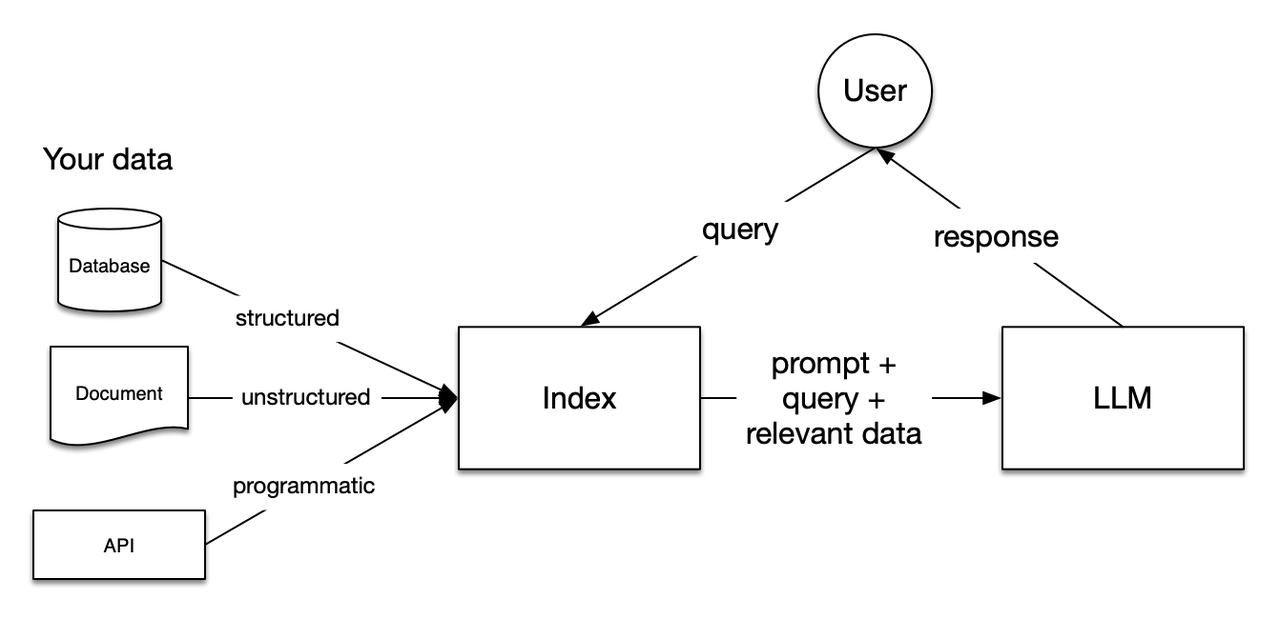
The general workflow:
Data Loading and Indexing:
- Load Data: Load private data (e.g., documents (unstructured), databases (structured), APIs).
- Create Index: Preprocess and index the loaded data for quick retrieval. The index is a structured intermediate representation that efficiently filters content relevant to queries.
User Query:
- Query the pre-created index first.
- Retrieval: Filter the most relevant content from the index.
- The retrieved relevant content forms the context used to assist the LLM’s generation process.
Response Generation:
- Combine Context and Query: Pass the retrieved relevant content (context) along with the user query to the LLM.
- Generate Response: The LLM uses this context to generate more accurate and relevant answers.

Technically, there are five stages:
Loading:
- Nodes and Documents: A
Documentis a container, encapsulating complete data source content, such as PDFs or APIs. ANodeis the atomic data unit in LlamaIndex, representing a “chunk” or fragment of a source Document, with its own metadata to link it to the document and other nodes. - Connectors: Also known as
Readers, process and convert data sources intoDocumentsandNodes.
- Nodes and Documents: A
Indexing:
- Indexes: Organized data indexes, e.g., stored as vector embeddings in a
VectorStore. The index also contains necessary metadata. - Embeddings: Numerical representations of data. These high-dimensional vectors capture semantic information, with semantically similar data being close in vector space, facilitating querying.
- Indexes: Organized data indexes, e.g., stored as vector embeddings in a
Storing: Storing the constructed indexes and other metadata to avoid repeated building.
Querying:
- Retrievers: Define how to efficiently retrieve relevant context from the index upon receiving a query. The retrieval strategy directly affects the relevance and efficiency of the retrieved data.
- Routers: Decide which retriever to use for retrieving relevant context from the knowledge base. Specifically, the
RouterRetrieverclass selects one or more candidate retrievers to perform the query, with aselectordeciding the best retriever based on metadata and query content. - Node Postprocessors: Apply transformations, filtering, or reordering logic to a set of retrieved nodes.
- Response Synthesizers: Concatenate the user query with retrieved context and prompts, generating responses based on the large model.
Evaluation: Assess the accuracy of query strategies, pipelines, and results.
3. Llama index Usage Example
We used ollama to deploy a 7B llama3 in the could, with documents from a short text (78KB) and ran the following code on a Mac (Core i7 2.6 GHz).
import time
from llama_index.core import VectorStoreIndex, SimpleDirectoryReader, Settings
from llama_index.embeddings.huggingface import HuggingFaceEmbedding
from llama_index.llms.ollama import Ollama
start_time = time.time()
load_start = time.time()
documents = SimpleDirectoryReader("data").load_data()
load_end = time.time()
embed_start = time.time()
Settings.embed_model = HuggingFaceEmbedding(model_name="BAAI/bge-base-en-v1.5")
embed_end = time.time()
llm_start = time.time()
Settings.llm = Ollama(model="llama3", request_timeout=360.0)
llm_end = time.time()
index_start = time.time()
index = VectorStoreIndex.from_documents(documents)
index_end = time.time()
query_engine_start = time.time()
query_engine = index.as_query_engine()
query_engine_end = time.time()
query_start = time.time()
response = query_engine.query("What did the author do growing up?")
query_end = time.time()
print(response)
print(f"Data loading time: {load_end - load_start} seconds")
print(f"Embedding model setup time: {embed_end - embed_start} seconds")
print(f"LLM setup time: {llm_end - llm_start} seconds")
print(f"Index creation time: {index_end - index_start} seconds")
print(f"Query engine creation time: {query_engine_end - query_engine_start} seconds")
print(f"Query execution time: {query_end - query_start} seconds")
print(f"Total time: {time.time() - start_time} seconds")输出:
According to the provided context, before college, the author worked on writing and programming outside of school. Specifically, he wrote short stories in his teenage years and tried writing programs on an IBM 1401 computer using an early version of Fortran in 9th grade (when he was around 13 or 14).
Data loading time: 0.021808862686157227 seconds
Embedding model setup time: 3.6557559967041016 seconds
LLM setup time: 0.0005099773406982422 seconds
Index creation time: 10.546114921569824 seconds
Query engine creation time: 0.0671701431274414 seconds
Query execution time: 1.3822910785675049 seconds
Total time: 15.673884868621826 secondsEven for a 78KB document, creating the index and querying took about 15 seconds, with over 10 seconds spent on index creation. We will analyze the reason for this time overhead in the following sections.
4. Llama index Source Code Analysis
4.1 Loading
Loading mainly has three modes: reading from files, reading from databases, and directly constructing document objects from text.
We will explain using SimpleDirectoryReader and DatabaseReader as examples.
It is worth noting that the llama_hub ecosystem provides many reader options.
4.1.1 SimpleDirectoryReader
SimpleDirectoryReader reads from a directory, constructing a document for each file.
from llama_index.core import SimpleDirectoryReader
documents = SimpleDirectoryReader("./data").load_data()Key path: load_data(), supports num_workers for multi-process reading (process pool).
# llama-index-core/llama_index/core/readers/file/base.py
class SimpleDirectoryReader(BaseReader, ResourcesReaderMixin, FileSystemReaderMixin):
def load_data(
self, # ...
) -> List[Document]:
# ...
if num_workers and num_workers > 1:
# multiprocessing ...
else:
# ...
for input_file in files_to_process:
documents.extend(
SimpleDirectoryReader.load_file(
# ...
)
)
return self._exclude_metadata(documents)In the load_file() method, there are two paths: for special files like ['.pdf', '.docx', '.pptx', '.png', '.mp3', '.mp4', '.csv', '.md', '.mbox', '.ipynb'], there are default cls readers; otherwise, it directly uses fs.open() to read text.
# llama-index-core/llama_index/core/readers/file/base.py
class SimpleDirectoryReader(BaseReader, ResourcesReaderMixin, FileSystemReaderMixin):
@staticmethod
def load_file(
input_file: Path, # ...
) -> List[Document]:
# ...
if file_suffix in default_file_reader_suffix or file_suffix in file_extractor:
# specific files ...
documents.extend(docs)
else:
# common text file ...
with fs.open(input_file, errors=errors, encoding=encoding) as f:
data = f.read().decode(encoding, errors=errors)
doc = Document(text=data, metadata=metadata or {})
documents.append(doc)
return documents4.1.2 DatabaseReader
DatabaseReader reads from a database, requiring users to write SQL. Additionally, this is actually a plugin.
from llama_index.readers.database import DatabaseReader
connection_uri = "sqlite:///example.db"
reader = DatabaseReader(uri=connection_uri)
query = "SELECT * FROM users"
documents = reader.load_data(query=query)
# llama-index-readers-database/llama_index/readers/database/base.py
class DatabaseReader(BaseReader):
def load_data(self, query: str) -> List[Document]:
with self.sql_database.engine.connect() as connection:
# ...
result = connection.execute(text(query))
for item in result.fetchall():
doc_str = ", ".join(
[f"{col}: {entry}" for col, entry in zip(result.keys(), item)]
)
documents.append(Document(text=doc_str))
return documentsThis reader directly uses the user query and fetchall to return all items, then encapsulates the text into Document.
4.2 Transformation
After reading data into documents, we need to perform transformations such as chunking, extracting metadata, embedding, etc. The input and output of transformations are Nodes (note that a document is a subclass of Node).
llama_index provides both high-level and low-level APIs, giving users a flexible range of options.
4.2.1 NodeParser
NodeParsers have three main types: File-Based Node Parsers, Text-Splitters, and Relation-Based Node Parsers. They take nodes as input (a document is also a node) and output processed nodes, commonly used for transformation.
For example, a file-based node parser might be used to transform nodes derived from file data.
from llama_index.core.node_parser import SimpleFileNodeParser
from llama_index.readers.file import FlatReader
from pathlib import Path
md_docs = FlatReader().load_data(Path("./test.md"))
parser = SimpleFileNodeParser()
md_nodes = parser.get_nodes_from_documents(md_docs)Text-Splitters will be discussed in more detail later.
Relation-Based Node Parsers currently include only the HierarchicalNodeParser, which splits nodes into those with hierarchical relationships. For instance:
from llama_index.core.schema import Document
from llama_index.core.node_parser import (
HierarchicalNodeParser,
get_leaf_nodes,
get_root_nodes,
)
doc_text = """ ... """
docs = [Document(text=doc_text)]
# default chunk size [2048, 512, 128]
node_parser = HierarchicalNodeParser.from_defaults()
nodes = node_parser.get_nodes_from_documents(docs)
# Get specific kind of nodes
leaf_nodes = get_leaf_nodes(nodes)
root_nodes = get_root_nodes(nodes)
level_nodes = get_deeper_nodes(nodes, depth=2)The root node instance:
TextNode(
id_='6562f0e3-e0bf-47c3-a75f-df3f19b99e71',
embedding=None,
metadata={},
excluded_embed_metadata_keys=[],
excluded_llm_metadata_keys=[],
relationships={
<NodeRelationship.SOURCE: '1'>: RelatedNodeInfo(
node_id='8001ac4d-f9b0-4b51-b3e4-1f61aa44449a',
# ...
),
<NodeRelationship.CHILD: '5'>: [
RelatedNodeInfo(
node_id='921ce06b-7880-483c-bf00-3ebf4746dc83',
# ...
)
]
},
text='...',
# ...
)And a leaf node instance:
TextNode(
id_='2ab92648-63d2-4197-98e5-b9afb892538d',
# ...
relationships={
<NodeRelationship.SOURCE: '1'>: RelatedNodeInfo(
node_id='921ce06b-7880-483c-bf00-3ebf4746dc83',
# ...
),
<NodeRelationship.PARENT: '4'>: RelatedNodeInfo(
node_id='921ce06b-7880-483c-bf00-3ebf4746dc83',
# ...
)
},
)To access nodes at a specific level after splitting nodes with inheritance relationships, llama_index uses a traversal strategy, which is not so efficient.
# llama-index-core/llama_index/core/node_parser/relational/hierarchical.py
def get_deeper_nodes(nodes: List[BaseNode], depth: int = 1) -> List[BaseNode]:
"""Get children of root nodes in given nodes that have given depth."""
# ...
root_nodes = get_root_nodes(nodes)
deeper_nodes = root_nodes
for _ in range(depth):
deeper_nodes = get_child_nodes(deeper_nodes, nodes)
return deeper_nodes
def get_root_nodes(nodes: List[BaseNode]) -> List[BaseNode]:
root_nodes = []
for node in nodes:
if NodeRelationship.PARENT not in node.relationships:
root_nodes.append(node)
return root_nodes
def get_child_nodes(nodes: List[BaseNode], all_nodes: List[BaseNode]) -> List[BaseNode]:
children_ids = []
for node in nodes:
if NodeRelationship.CHILD not in node.relationships:
continue
children_ids.extend(
[r.node_id for r in node.relationships[NodeRelationship.CHILD]]
)
child_nodes = []
for candidate_node in all_nodes:
if candidate_node.node_id not in children_ids:
continue
child_nodes.append(candidate_node)
return child_nodesDue to the fact that HierarchicalNodeParser returns all nodes in a single list, we must retrieve nodes from a specific layer (using get_deeper_nodes) before constructing the index and executing embedding. Otherwise, the nodes will contain a lot of redundant content, significantly reducing the overall efficiency of RAG.
This inheritance relationship information can be used with the AutoMergingRetriever, as shown in auto_merger.
This involves sending leaf nodes into the index construction process but merging leaf nodes automatically to obtain richer context, leading to higher final scores.
| Names | Correctness | Relevancy | Faithfulness | Semantic Similarity |
|---|---|---|---|---|
| Auto Merging Retriever | 4.266667 | 0.916667 | 0.95 | 0.962196 |
| Base Retriever | 4.208333 | 0.916667 | 0.95 | 0.960602 |
4.2.2 High level api:VectorStoreIndex
from llama_index.core import VectorStoreIndex, SimpleDirectoryReader
documents = SimpleDirectoryReader("./data").load_data()
vector_index = VectorStoreIndex.from_documents(documents)
vector_index.as_query_engine()We build index from documents:
# llama_index/core/indices/base.py
class BaseIndex(Generic[IS], ABC):
@classmethod
def from_documents(
cls: Type[IndexType], documents: Sequence[Document], # ...
) -> IndexType:
# ...
nodes = run_transformations(
documents, # type: ignore
transformations,
# ...
)
return cls(
nodes=nodes,
# ...
)
# llama_index/core/ingestion/pipeline.py
def run_transformations(
nodes: List[BaseNode], transformations: Sequence[TransformComponent], # ...
) -> List[BaseNode]:
if not in_place:
nodes = list(nodes)
for transform in transformations:
if cache is not None:
hash = get_transformation_hash(nodes, transform)
cached_nodes = cache.get(hash, collection=cache_collection)
if cached_nodes is not None:
nodes = cached_nodes
else:
nodes = transform(nodes, **kwargs)
cache.put(hash, nodes, collection=cache_collection)
else:
nodes = transform(nodes, **kwargs)
return nodesIf the user does not explicitly pass them in, the default transformation is SentenceSplitter, which has a long subclass inheritance chain:
Representation -> BaseModel -> BaseComponent -> TransformComponent -> NodeParser -> TextSplitter -> MetadataAwareTextSplitter -> SentenceSplitter
Calling transform() leads to the base class’s __call__ method, which processes down to:
# llama_index/core/node_parser/text/sentence.py
class SentenceSplitter(MetadataAwareTextSplitter):
def _split_text(self, text: str, chunk_size: int) -> List[str]:
# ...
splits = self._split(text, chunk_size)
chunks = self._merge(splits, chunk_size)
return chunks
def _split(self, text: str, chunk_size: int) -> List[_Split]:
token_size = self._token_size(text)
if token_size <= chunk_size:
return [_Split(text, is_sentence=True, token_size=token_size)]
text_splits_by_fns, is_sentence = self._get_splits_by_fns(text)
text_splits = []
for text_split_by_fns in text_splits_by_fns:
token_size = self._token_size(text_split_by_fns)
if token_size <= chunk_size:
text_splits.append(
_Split(
text_split_by_fns,
is_sentence=is_sentence,
token_size=token_size,
)
)
else:
recursive_text_splits = self._split(
text_split_by_fns, chunk_size=chunk_size
)
text_splits.extend(recursive_text_splits)
return text_splitsHere, _split splits the text into multiple _Split blocks. The splitting strategy follows this sequence:
- Split by paragraphs (
'\n\n\n'). - Split by the NLTK sentence tokenizer (a pip package based on the Punkt model).
- Split using a regex
[^,\.;]+[,\.;]?(a regular expression that captures strings + punctuation (comma, period, semicolon)). - Split by
" "(space). - Split by character(Nearly never used).
If the chunk size is satisfied, it returns early without further splitting. Note the recursive logic recursive_text_splits.
For example, our sample text, with 75,000 characters, will be split into a list with len(splits)=759.
_merge then merges the splits into formal chunks.
# llama_index/core/node_parser/text/sentence.py
class SentenceSplitter(MetadataAwareTextSplitter):
def _merge(self, splits: List[_Split], chunk_size: int) -> List[str]:
# ...
def close_chunk() -> None:
# finish a chunk and then create a new one
pass
while len(splits) > 0:
cur_split = splits[0]
if cur_split.token_size > chunk_size:
raise ValueError("Single token exceeded chunk size")
if cur_chunk_len + cur_split.token_size > chunk_size and not new_chunk:
# if adding split to current chunk exceeds chunk size
close_chunk()
else:
if (
cur_split.is_sentence
or cur_chunk_len + cur_split.token_size <= chunk_size
or new_chunk # new chunk, always add at least one split
):
# add split to chunk
cur_chunk_len += cur_split.token_size
cur_chunk.append((cur_split.text, cur_split.token_size))
splits.pop(0)
new_chunk = False
else:
# close out chunk
close_chunk()
# handle the last chunk
if not new_chunk:
chunk = "".join([text for text, length in cur_chunk])
chunks.append(chunk)
# run postprocessing to remove blank spaces
return self._postprocess_chunks(chunks)If the chunk size condition is met, it merges as many splits as possible into one chunk. Note there is chunk overlap, where adjacent chunks overlap some text (default 200 tokens), providing better contextual continuity.
For our example document, after merging into chunks, len(chunks)=22.
After _split and _merge, chunks are encapsulated into nodes and returned.
Performance-wise, this part involves complex Python processing, with O(N) linear growth, which could be optimized using C++/parallelism (available in the low-level API). Additionally, logical optimization is possible (splits are too fine and chunks merged with many cycles, as well as meaningless pop(0)). There’s also overhead in initializing the tokenizer and making multiple tokenize() calls
4.2.3 Low level api:IngestionPipeline
from llama_index.core import SimpleDirectoryReader, VectorStoreIndex
from llama_index.core.ingestion import IngestionPipeline
from llama_index.core.node_parser import TokenTextSplitter
documents = SimpleDirectoryReader("./data").load_data()
pipeline = IngestionPipeline(transformations=[TokenTextSplitter(), embed_model, ...])
nodes = pipeline.run(documents=documents)
index = VectorStoreIndex(nodes)The pipeline’s run method supports parallelism and caching.
# llama_index/core/ingestion/pipeline.py
class IngestionPipeline(BaseModel):
def run(
self, # ...
) -> Sequence[BaseNode]:
input_nodes = self._prepare_inputs(documents, nodes)
# ...
if num_workers and num_workers > 1:
# multiprocessing ...
else:
nodes = run_transformations(
nodes_to_run,
self.transformations,
cache=self.cache if not self.disable_cache else None,
# ...
)
if self.vector_store is not None:
self.vector_store.add([n for n in nodes if n.embedding is not None])
return nodescache is applied during run_transformations, using nodes and transform hash values as keys for storage, avoiding repeated computations in the pipeline for repetitive tasks.
For IngestionCache, the applied cache is essentially SimpleCache (an in-memory cache).
llama_index also supports other DB caches, such as those based on sqlalchemy for database integration.
4.3 Indexing & Embedding
4.3.1 Basic Concepts
After constructing nodes through transformation, we need to process these nodes into an index (a data structure for querying). This step is also where document processing consumes the most time. We will illustrate this using VectorStoreIndex as an example.
VectorStoreIndex creates a vector embedding for each node. An embedding is a numerical representation of text data, where semantically similar texts have similar embeddings. This allows for semantic search instead of simple keyword matching for queries.
For example, two sentences with the same meaning will have a high cosine similarity (essentially, the cosine of the angle between the vectors, where 1 indicates the same direction).
Embedding for 'The cat is on the mat.':
[ 0.021, 0.012, -0.034, 0.045, 0.038, -0.026, 0.056, -0.024, 0.013, -0.017]
Embedding for 'The feline is on the rug.':
[ 0.023, 0.010, -0.032, 0.046, 0.036, -0.025, 0.057, -0.022, 0.011, -0.018]
Cosine Similarity(余弦相似度): 0.995During querying, the query is also converted to an embedding, and then a similarity calculation is performed with all nodes. The top-k most similar embeddings are returned.
Other types of indexes include:
SummaryIndex(formerly List Index)
TreeIndex: Nodes have a tree-like storage structure (inheritance relationship)
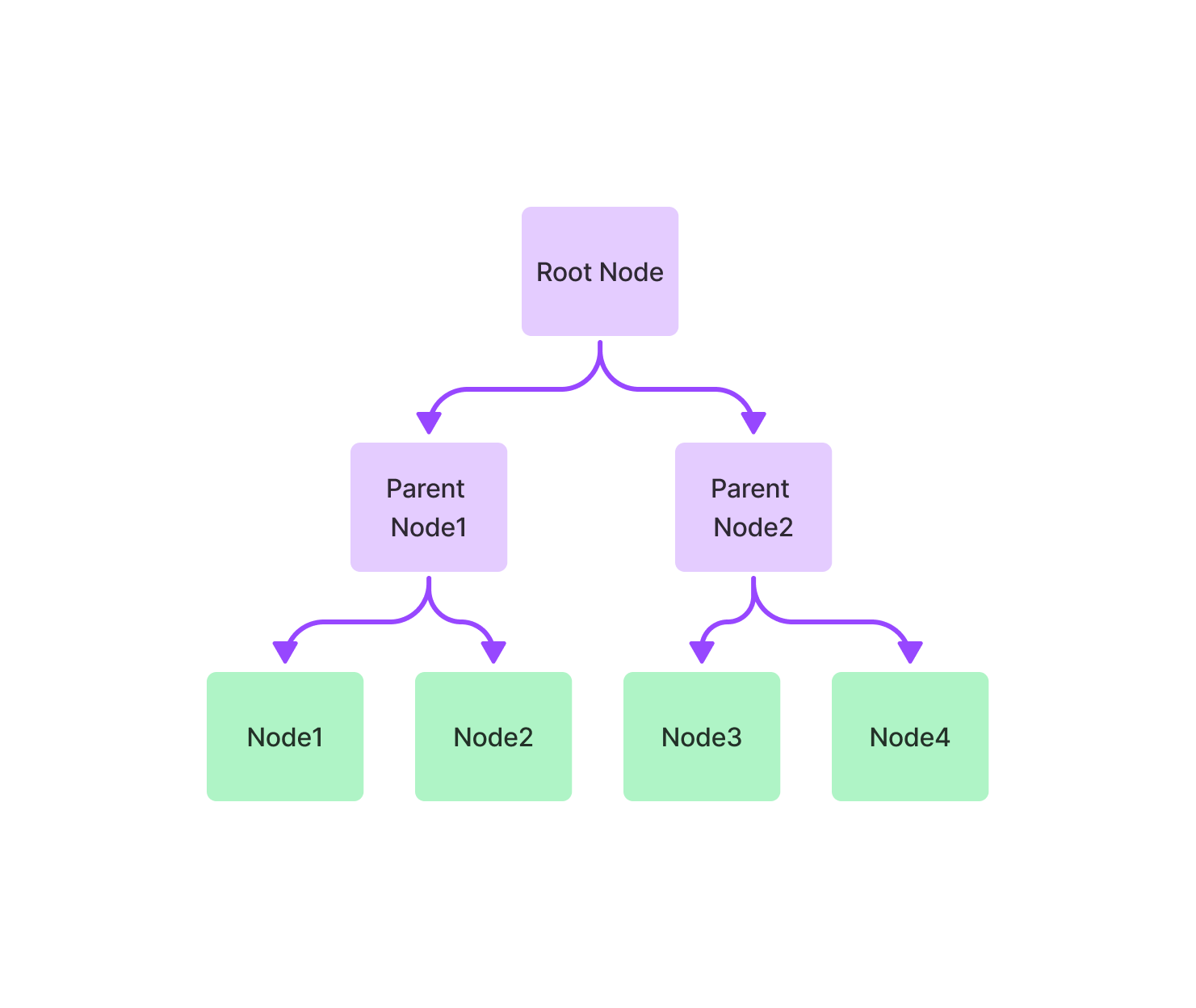 Tree nodes facilitate the retrieval of nodes with inheritance relationships, starting from the root and querying down to the leaf node.
Tree nodes facilitate the retrieval of nodes with inheritance relationships, starting from the root and querying down to the leaf node.Keyword Table Index
property graph index: for instance
+---------+ +---------+
| | WorksAt | |
| Alice +------------->| OpenAI |
| | | |
+----+----+ +---------+
|
| Knows
|
+----v----+
| |
| Bob |
| |
+----+----+
|
| WorksAt
|
+----v----+
| |
|Microsoft|
| |
+---------+4.3.2 Process of building index
# llama_index/core/indices/vector_store/base.py
class VectorStoreIndex(BaseIndex[IndexDict]):
def __init__(
self, nodes: Optional[Sequence[BaseNode]] = None, # ...
) -> None:
# ...
super().__init__(
nodes=nodes, index_struct=index_struct, # ...
)
# llama_index/core/indices/base.py
class BaseIndex(Generic[IS], ABC):
def __init__(
self, nodes: Optional[Sequence[BaseNode]] = None, # ...
) -> None:
# ...
if index_struct is None:
nodes = nodes or []
index_struct = self.build_index_from_nodes(
nodes + objects # type: ignore
)
self._index_struct = index_struct
self._storage_context.index_store.add_index_struct(self._index_struct)
self._transformations = (
transformations
or transformations_from_settings_or_context(Settings, service_context)
)During base class initialization, build_index_from_nodes is called, which eventually calls _add_nodes_to_index, and then processes each node through get_text_embedding_batch.
# llama_index/core/indices/vector_store/base.py
class VectorStoreIndex(BaseIndex[IndexDict]):
def _add_nodes_to_index(
self, index_struct: IndexDict, nodes: Sequence[BaseNode], # ...
) -> None:
# ...
for nodes_batch in iter_batch(nodes, self._insert_batch_size):
nodes_batch = self._get_node_with_embedding(nodes_batch, show_progress)
new_ids = self._vector_store.add(nodes_batch, **insert_kwargs)
if not self._vector_store.stores_text or self._store_nodes_override:
for node, new_id in zip(nodes_batch, new_ids):
# NOTE: remove embedding from node to avoid duplication
node_without_embedding = node.copy()
node_without_embedding.embedding = None
index_struct.add_node(node_without_embedding, text_id=new_id)
self._docstore.add_documents(
[node_without_embedding], allow_update=True
)
else:
# image embedding ...
# llama_index/core/indices/utils.py
def embed_nodes(
nodes: Sequence[BaseNode], embed_model: BaseEmbedding, # ...
) -> Dict[str, List[float]]:
# ...
new_embeddings = embed_model.get_text_embedding_batch(
texts_to_embed, show_progress=show_progress
)
for new_id, text_embedding in zip(ids_to_embed, new_embeddings):
id_to_embed_map[new_id] = text_embedding
return id_to_embed_mapWithin this call chain, we find a function wrapper mechanism @dispatcher.span. By inserting span before and after function calls, it captures and records each function’s execution time, inputs, outputs, and errors.
# llama_index/core/base/embeddings/base.py
class BaseEmbedding(TransformComponent):
@dispatcher.span
def get_text_embedding_batch(
self, texts: List[str], # ...
) -> List[Embedding]:
# ...
model_dict = self.to_dict()
for idx, text in queue_with_progress:
cur_batch.append(text)
if idx == len(texts) - 1 or len(cur_batch) == self.embed_batch_size:
# ...
embeddings = self._get_text_embeddings(cur_batch)
result_embeddings.extend(embeddings)
# ...
cur_batch = []
return result_embeddingsThere is a batch logic here, accumulating (default 10) texts before sending them for embedding.
Finally, it calls the model’s encode interface, such as the BAAI/bge-base-en-v1.5 embedding model, reaching the hugging face model embedding.
# llama_index/embeddings/huggingface/base.py
class HuggingFaceEmbedding(BaseEmbedding):
# ...
def _embed(
self, sentences: List[str], prompt_name: Optional[str] = None,
) -> List[List[float]]:
return self._model.encode(
sentences, batch_size=self.embed_batch_size, # ...
).tolist()Further calls lead to the encode interface of sentence_transformers/SentenceTransformer.py, which we won’t expand on here.
Returning along the call chain, nodes are added to index_struct, and node_without_embedding is added to self._docstore. This completes the indexing construction.
From a performance perspective, a simple time analysis shows that most time consumption comes from embedding construction (simulating a host scenario without GPU). This part has limited optimization potential. Currently, llama_index does not handle embedding parallelism well; it must be placed in the transformation phase (once inside the vector index initialization, it becomes irrelevant to parallelism).
_get_text_embeddings time: 5.118131160736084
_get_text_embeddings time: 2.813439130783081
all index time: 8.08009123802185